Viscoelastic enzyme
Nano-rheology of molecules: Different materials can be grouped according to their mechanical properties. For example, when subjected to a small shear force, solids respond with a deformation (strain) proportional to the force, while liquids respond with a flow (strain rate) proportional to the force. Enzymes do both.
Enzymes are very large molecules consisting of thousands of atoms. They are mesoscopic systems and one can investigate their mechanical properties. With the experiment described below, we discovered that enzymes are viscoelastic. It means the dynamical response is elastic at high frequencies and resembles a viscous flow at low frequencies.
While this is true of most materials in an extended frequency range (liquids have a shear modulus at high enough frequencies, typically above GHz; solids flow on sufficiently long timescales), for enzymes the frequency range separating elastic from viscous behavior is of the same order as the (inverse) time scales for large conformational motion of the molecule. In other words, it appears that visco-elasticity is fundamental for enzyme conformational dynamics.
The visco-elastic transition was discovered in the enzyme Guanylate Kinase (GK) by a simple experiment where we apply an oscillatory stress to the enzyme molecule and measure the amplitude of the resulting strain, with sub-Angstrom resolution. We refer to the method as “nano-rheology”. It works as follows.
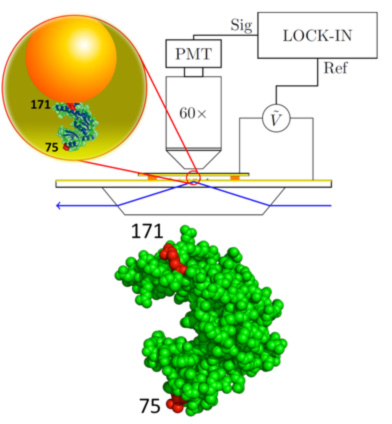
Figure 1 at left: Nano-rheology setup. Also shown is the structure of the enzyme Guanylate Kinase (GK).
The enzyme under study tethers 20 nm gold nanoparticles (GNPs) to the gold-coated surface of a glass slide. Separated by a 200 μm gap, a similarly coated cover slip forms the second electrode of this parallel plate capacitor (Fig. 1). The enzyme is specifically anchored to both the slide and GNP through the gold-sulfur bond due to cysteins introduced by mutagenesis on diametrically opposite sides of the molecule. The sample is in buffer and generally physiological conditions.
A ∼ 500 mV oscillating voltage applied at the electrodes drives the negatively charged GNPs, resulting in an oscillatory stress on the molecules. The amplitude of the oscillatory motion of the GNPs, reporting on the strain of the enzymes, is measured by evanescent wave scattering in a phase-locked loop, ensemble averaged over ∼108 GNPs.
The nm-scale thermal motion of individual GNPs ensemble averages to zero, allowing the imposed collective oscillation to be measured with sub-Å resolution. In the apparatus the enzyme is in its folded, functional state.
Figure 2 shows the viscoelastic mechanical response of the enzyme molecule of Fig. 1 (GK). The amplitude of the strain "diverges" at low frequency: the signature of viscoelasticity.
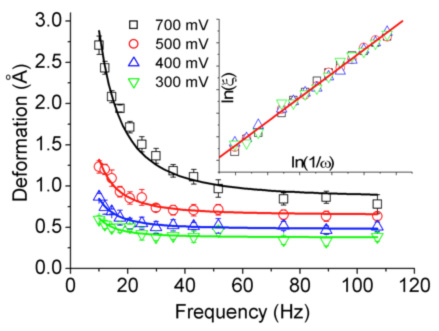
Figure 2 at right: rheological response of the enzyme GK: amplitude of the molecule’s deformation vs frequency of the applied force. The different curves correspond to different amplitudes of the applied force. The fits and the inset show that the data follow the Maxwell model of viscoelasticity.
Nano-rheology with surface plasmon resonance coupled optics: in the nano-rheology setup of Fig. 1 one can improve the optical detection by taking advantage of so-called “plasmon resonances”: electron resonances at optical frequencies in the gold layer and gold nanoparticles of the setup.
Nano-rheology as a biochemical assay: nano-rheology provides a method for determining binding constants of ligands to enzymes in a label free assay. This method is orthogonal to traditional spectroscopic methods. Namely, ligand binding is detected through the resulting change in mechanical stiffness of the enzyme. Fig. 3 shows a binding isotherm obtained with this method, quantifying binding of the substrate GMP to the enzyme GK.
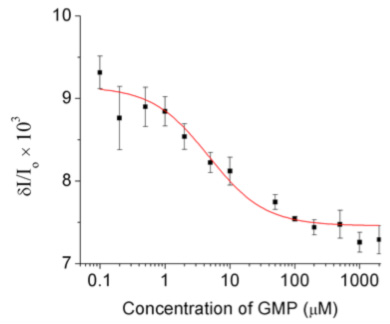
Figure 3 at left: binding curve for GMP binding to the enzyme GK, obtained with the nano-rheology method. The signal on the ordinate is the modulation of the scattered intensity, proportional to the amplitude of the deformation of the enzyme elicited by the applied oscillatory force.
In general, we think the nano-rheology approach is opening a new window on the dynamics of conformational changes in enzymes.
Associated publications:
- Y. Wang and G. Zocchi, "Elasticity of globular proteins measured from the AC susceptibility," Phys. Rev. Lett. 105, 238104 (2010).
- Y. Wang and G. Zocchi, "The folded protein as a visco-elastic solid," Europhys. Lett. 96, 18003 (2011).
- Y. Wang and G. Zocchi, "Viscoelastic transition and yield strain of the folded protein," PLoS ONE 6(12), e28097 (2011).
- H. Qu, J. Landy, and G. Zocchi, "Cracking phase diagram for the dynamics of an enzyme," Phys. Rev. E 86, 041915 (2012).
- A. Ariyaratne, C. Wu, C-Y. Tseng, and G. Zocchi, "Dissipative Dynamics of Enzymes," Phys. Rev. Lett. 113, 198101 (2014).
- A. Ariyaratne and G. Zocchi, "Plasmon resonance enhanced mechanical detection of ligand binding," Appl. Phys. Lett. 106, 013702 (2015).