Giovanni Zocchi
Molecular Biophysics
Office: 3-124 Knudsen ; (310) 825-4018
Lab: A-425, A-429 PAB ; (310) 206-0715 zocchi@physics.ucla.edu

Education:
- Ph.D. in Physics, University of Chicago (1990)
- Undergraduate degree (Laurea) in Physics, Universita’ di Pisa and Scuola Normale Superiore, Pisa, Italy (1985)
Appointments:
- 2005 - pres. Associate Professor of Physics, UCLA
- 1999 - 2005 Assistant Professor of Physics, UCLA
- 1993 - 1999 Research Faculty, Niels Bohr Institute, Copenhagen (Denmark)
- 1990 - 1993 Postdoctoral Fellow, Ecole Normale Superieure, Paris (France)
- 1985 - 1990 Research Associate, University of Chicago
Past Research. My Ph.D. and postdoctoral research was in the field of non-linear dynamics, instabilities, and turbulence. For a brief description, click here.
Present Research. The main research topic in the lab is the study of conformational changes of biological macromolecules (proteins, DNA). The ability of biological molecules to perform specific tasks through directed conformational motion is the molecular basis of life. Our goal is to understand the minimal design requirements for such molecular machines. Then we can build our own.
We have developed a single-molecule method which detects conformational changes of single biomolecules with 1 nm resolution (Fig. 1). It is a unique tool to study, for instance, the dynamics of molecular recognition events (Fig. 2).
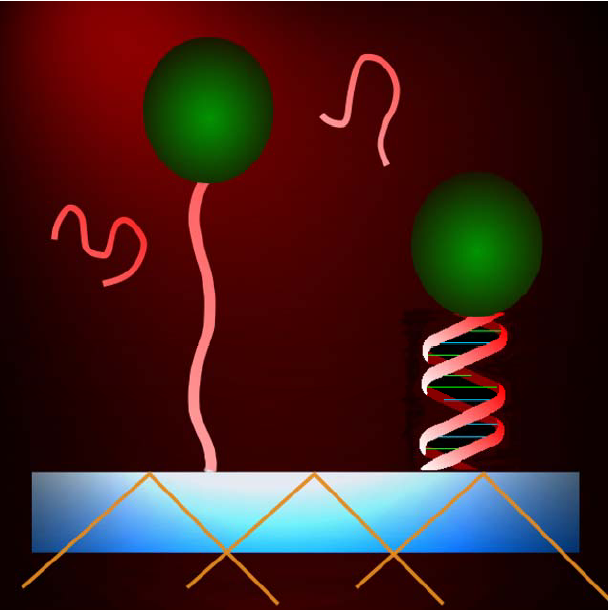
| Fig. 1. The micron size bead (not to scale) is tethered to the glass slide by a single 10 - 20 nm long DNA molecule. A conformational change of the DNA, in this case induced by binding of a complementary strand, displaces the bead with respect to the surface. The bead’s motion is detected with sub-nm resolution by evanescent wave scattering [Proc. Natl. Acad. Sci. USA 100, 7605-10 (2003)]. |
 |
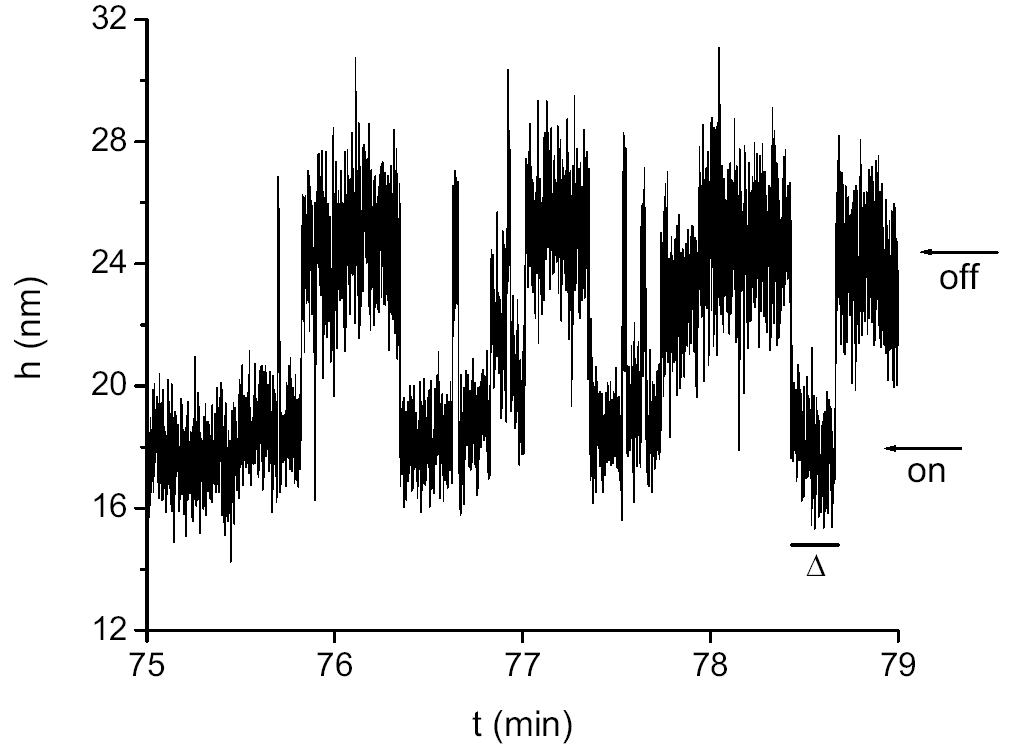 |
Fig. 2. This trace shows, in real time, a single Integration Host Factor (IHF) protein binding to (ON) and falling off from (OFF) a single 76 base pair long DNA molecule. When the protein binds, the DNA bends around it, so its end-to-end distance shortens (by about 7 nm); this conformational change is detected by the method of the previous figure [Phys. Rev. Lett. 94, 118101 (2005)]. |
| Force naturally couples to conformational motion. We have invented a mechanical approach to control the function of virtually any protein. We insert an externally controllable “molecular spring” on the protein: the tension of the spring controls the protein’s conformation and thus its function (Fig. 3). This approach opens a new window on the inner workings of proteins, specifically the central property of allostery (the ability of proteins to change conformation in response to a chemical signal). |
|
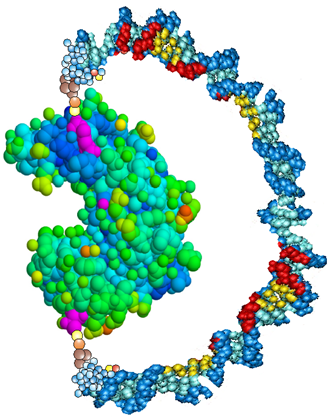
| Fig. 3. A “molecular spring” made of a 60 bases long piece of DNA is wound around the enzyme Guanylate Kinase. In the single-stranded form, the DNA is flexible and does not stress the protein. Hybridization with a complementary strand stiffens the molecular spring, which then exerts a mechanical stress on the protein, deforming the substrate binding pocket (i.e. inducing a conformational change). The activity of the enzyme can thus be externally controlled [Phys. Rev. Lett. 95, 078102 (2005)]. |
 |
Lab Website:
Zocchi Lab for Molecular Biophysics
Links:
Fire of Desire (http://wordsmitten.com/storycove_mukta.htm)
California Sailing Academy (http://www.californiasailingacademy.com/)
|



